 Using pROC
Using pROC
Now that pROC is released, it is time to see how it can be used. Here is an example of a typical analysis:
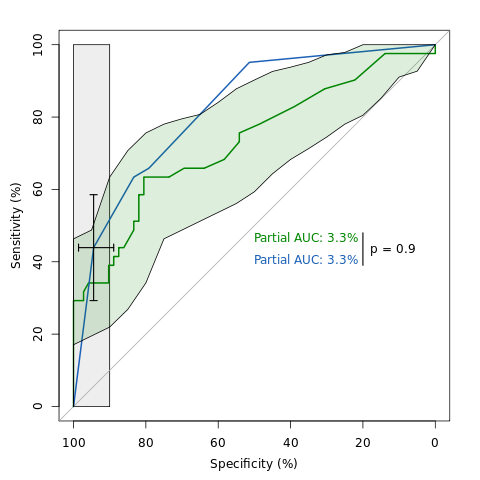 An example of ROC analysis with pROC.
An example of ROC analysis with pROC.
Here is the code that was used to generate this image. The first step is to load pROC (don't forget to type install.packages("pROC") if you haven't installed pROC yet!) and the aSAH data that is provided:
library(pROC) data(aSAH)
First we'll build the ROC curves. We want the sensitivity and specificity in % (thus percent = TRUE) and a partial AUC between 90 and 100% specificity:
rocs100 <- roc(aSAH$outcome, aSAH$s100, percent = TRUE, partial.auc=c(100, 90)) rocwfns <- roc(aSAH$outcome, aSAH$wfns, percent = TRUE, partial.auc=c(100, 90))
Then we compute confidence intervals. For S100, we compute the confidence of sensitivity over several levels of specificity. This will allow us to plot the interval as a shape. With WFNS, we compute the confidence interval of the cut-off 4.5. We use 10000 bootstrap replicates to get a good estimation:
cis100 <- ci(rocs100, of = "se", sp = seq(0, 100, 5), boot.n=10000) ciwfns <- ci(rocwfns, of = "threshold", threshold=4.5, boot.n=10000)
Now, the main feature of pROC is the comparison of two ROC curves. Again we use 10000 bootstrap replicates to get a good estimation:
rtest <- roc.test(rocs100, rocwfns, boot.n=10000)
And finally we plot the ROC curves:
png("demo.png")
plot(rocs100, col="#008600", max.auc.polygon=TRUE, print.auc=TRUE, print.auc.y=48)
plot(rocwfns, col="#1c61b6", add=TRUE, print.auc=TRUE, print.auc.y=42)
And the confidence intervals:
plot(cis100, type="shape", col="#00860022", no.roc=TRUE) plot(ciwfns, type="bars", lwd=1.5)
Last we add the p value of the ROC comparison test:
text(18, 45, sprintf("p = %.1f", rtest$p.value), adj=c(0,1))
lines(c(20, 20), c(39, 48), lwd=1.5)
dev.off()
And that's it, we have generated the image above!
You can find more such examples on the screenshots page on ExPASy!
Xavier Robin
Published Tuesday, April 27, 2010 20:50 CEST
Permalink: /blog/2010/04/27/using-proc
Tags:
pROC
Comments: 0
Comments
No comment
 Search
Search Tags
Tags Recent posts
Recent posts Calendar
Calendar Syndication
Syndication Recommend
Recommend